Recognizing antibiotic-resistant MRSA infections sooner is key to successful treatment

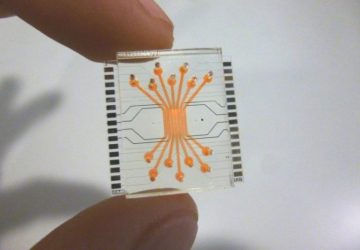
Researchers Anwar Sunna (left) and Vinoth Kumar Rajendran used a smartphone to make an MRSA detector.
Researchers in Australia have developed an inexpensive, portable device that can detect a deadly strain of bacteria in under 40 minutes.
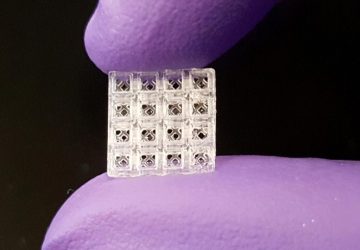
The device consists entirely of off-the-shelf and 3D-printed parts that cost a total of US $26, and the results can be read by an app on a smartphone. The developers of the device, at Macquarie University in Sydney, described their system last month in the journal Sensors and Actuators B: Chemical.
The group is using this diagnostic to go after a superbug known as MRSA (Methicillin-resistant Staphylococcus aureus), which strikes students, athletes, soldiers, hospital patients, and other communities in close quarters. MRSA has developed resistance to some of our best antibiotics and, if not successfully treated, can result in sepsis and death.
The key to fighting MRSA is to hit it with a specific antibiotic to which it has not yet developed resistance. But first doctors must test the infection to confirm it is the antibiotic-resistant kind.
Getting results from those tests can take anywhere from half a day to two days, depending on the type of test and the facility’s resources. Meanwhile, a patient’s condition can deteriorate rapidly.
The new device could cut diagnostic wait time down to less than an hour and provide an inexpensive solution in resource-poor settings.
“The idea is to be able to use it in remote areas where there may not be hospitals everywhere, or machines and technicians capable of doing this sort of test,” says Anwar Sunna, an associate professor in the department of molecular sciences at Macquarie University who supervised the research. He notes that the device will require a lot more work before it is ready for the field.
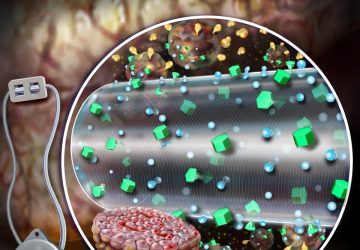
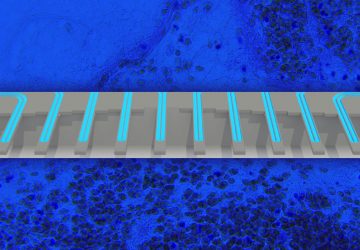
The Macquarie team’s technique identifies MRSA by looking for two key genes called femA and mecA. If the genes are present, quantum dots on test strips fluoresce, a smartphone camera registers the florescence through an attached filter, and an app displays the results.
Here’s how the system works, in more detail: First, DNA from a bacterium sample must be isolated and multiplied. The team approached this using a technique called convective polymerase chain reaction, or cPCR—a new take on traditional PCR, which is widely used, but is expensive and requires trained technicians. Sunna’s team 3D-printed a custom cPCR system.
Next the amplified DNA is subjected to a lateral flow assay. These test strips contain antibodies capable of capturing the femA and mecA sequences. Quantum dots on the test strip act as labels, fluorescing when the key DNA sequences are captured.
A phone is then placed in a holder that looks like a bulky mobile phone case. This phone holder includes an optical module with a filter, LED light, and a slot to insert the test strips. To read the sample, the device shines light on the strips, and if the quantum dots fluoresce, the phone camera picks it up with the help of the filter, and the results are displayed on an app.
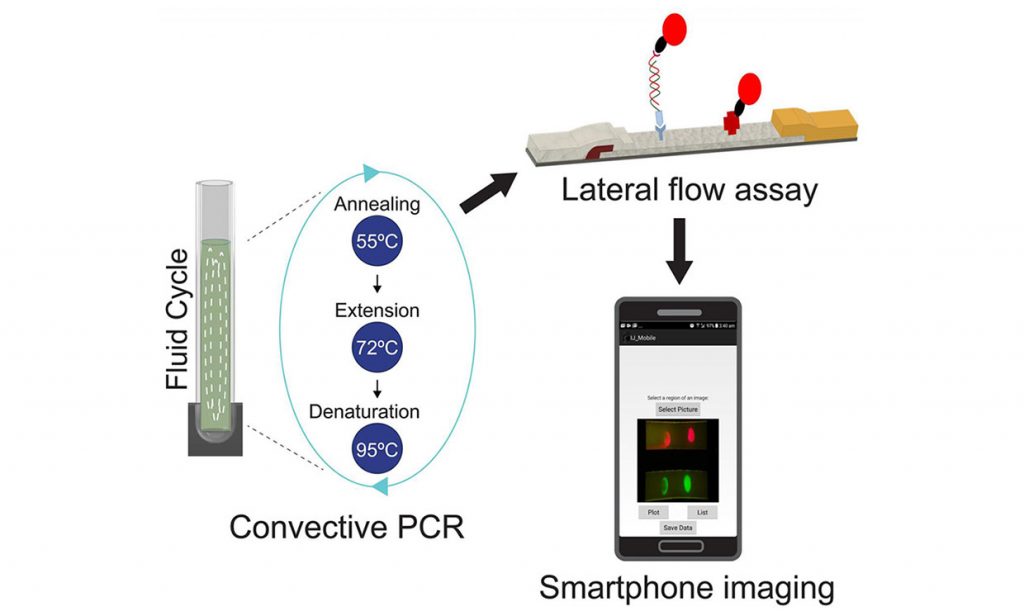
To search for MRSA, a new device subjects a DNA sample to a lateral flow assay where antibodies capture two genes from the bacterium. Quantum dots on the test strips fluoresce when those genes are attached, and results are shown in an app.
None of these techniques or assays are novel. For example, Ahram Biosystems sells a cPCR device that can detect some bacteria. It sells for about $4,000.
But the Macquarie group was able to combine existing techniques specifically for use on MRSA. Their system is built with inexpensive components that run on a battery and connect directly to a smartphone. The device is also an advance on previous designs in that it can detect more than one DNA sequence at a time, Sunna says.
A rapid, portable device would be a big change for many health care settings, which still rely on old-fashioned culturing, where the bacteria is allowed to grow on a plate of nutrients and checked after a day or two to find out what it is. “I was surprised when my student, who had gone to one of the big labs in Sydney, came back to me and said the test was still run using cultivation on plates,” says Sunna.
In some cases, a day or two is just too long to wait. Says Sunna: “Doctors will start treating you with antibiotics under the assumption you have an infection, but if you have an antibiotic-resistant organism, those antibiotics aren’t going to work. And by the time you get your answer, 24 hours later, if you have a blood infection, it becomes quite dangerous.”
source: www.spectrum.ieee.org