Engineered bacteria are being deployed to treat everything from autoimmune disease and metabolic disorder to diabetes and cancer. But once ingested or injected, it can be hard to know whether these “living medicines” are reaching their desired locations inside the body.
A new technique, described today in Nature, could allow clinicians to track therapeutic microbes with nothing more than an ultrasound detector—the same device already used routinely in medicine to view fetuses during pregnancy and for other diagnostic applications.
“This is an ingenious platform,” says James Collins, a synthetic biologist at MIT who was not involved in the study. “It’s a simple, beautiful design that has real value.”
Monitoring microbes that live deep inside our bodies is a big challenge. Existing methods either rely on fluorescent light (which can’t penetrate far below the skin’s surface), radiation (which can be hazardous to cells and has limited resolution), or magnetic resonance imaging (which requires large and expensive instruments).
A team from Caltech got around these problems by turning to sound waves—and by taking advantage of a natural quirk of aquatic microorganisms.
Many water-dwelling microbes have genes that encode gas-filled nanostructures to help them stay afloat. And as Caltech chemical engineer Mikhail Shapiro showed in 2014, these protein-encased shells—known as gas vesicles—reliably scatter high-frequency sound waves to generate an echo that can be converted via an ultrasound scanner into detailed images.
But that earlier work was done with purified gas vesicles from weird and wacky microbes, including one that thrives in salt mines and another found in brackish seas. Shapiro hadn’t yet shown it could work in living bacteria, let alone in those that have the ability to colonize the human body and could be suitable for therapeutic purposes.
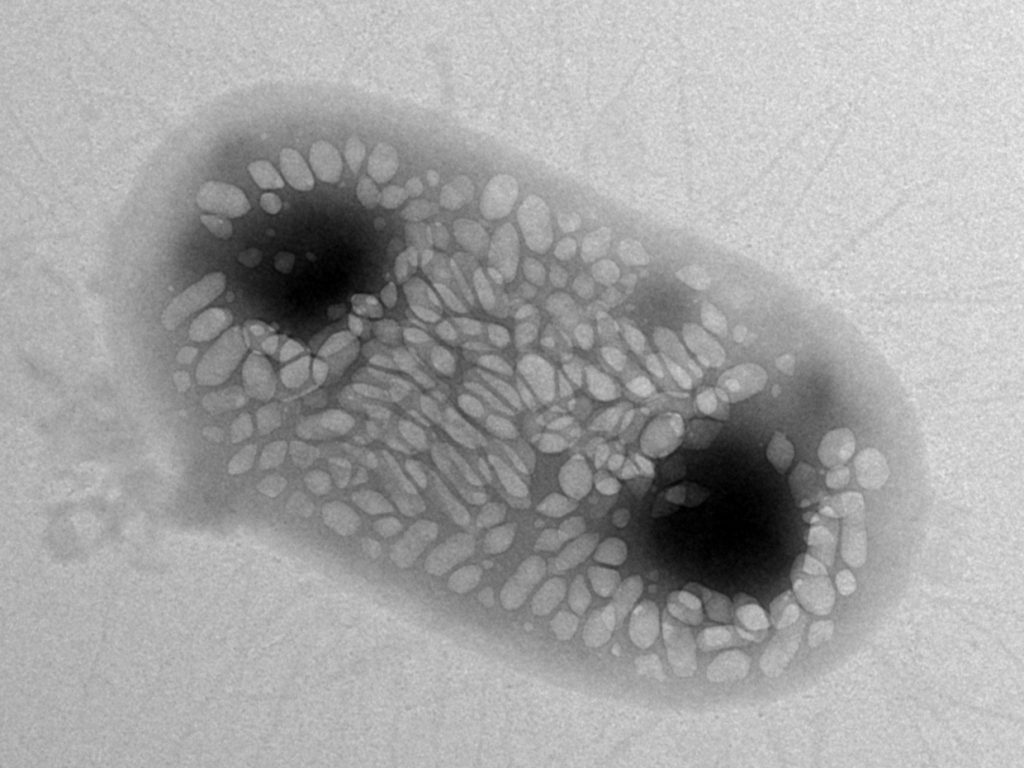
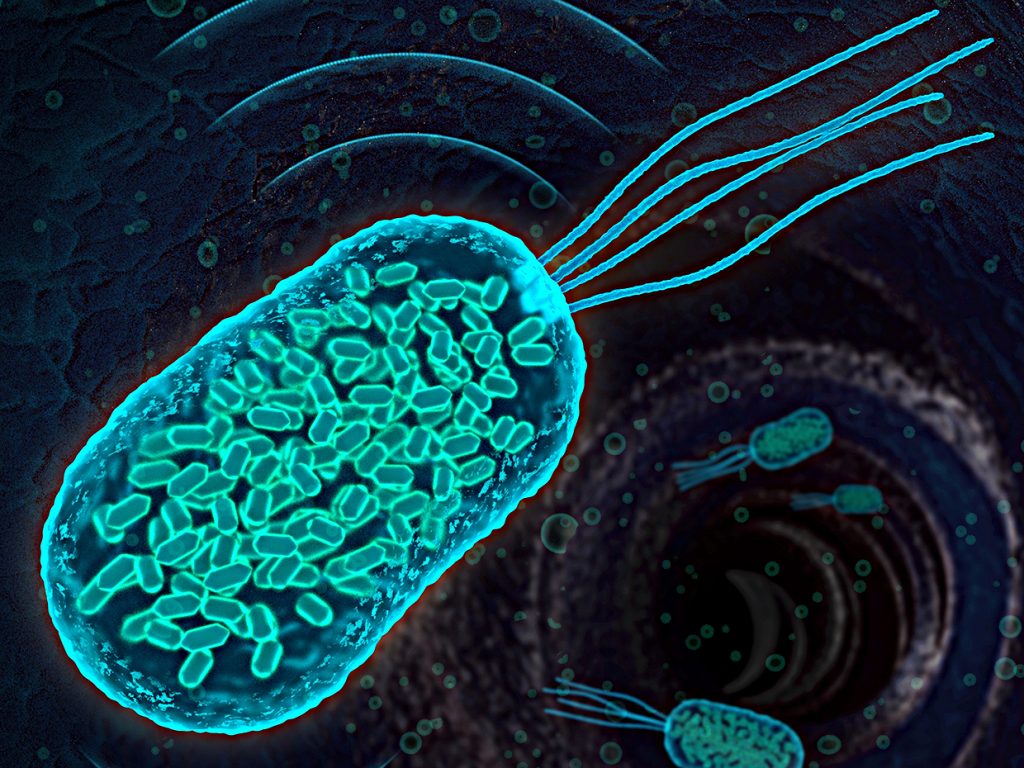
Image: Anupama Lakshmanan/CaltechThis image shows an E. coli bacterium that was genetically engineered to express gas vesicles in order to track its movement through a mouse’s body using ultrasound.
Shapiro and his colleagues have now engineered strains of the common gut bug Escherichia coli and the food-poisoning agent Salmonella typhimurium to do just that. They mixed and matched 11 different DNA components from two bacterial species—an aquatic plankton and a soil microbe, each of which naturally produce gas vesicles—to create what they term an ‘acoustic reporter gene.’
“By including these acoustic reporter genes into these engineered cellular constructs,” Shapiro says, “we now should be able to actually follow what’s happening with the therapy.”
To test that idea, the researchers inserted the gene cluster into E. coli or S. typhimurium so that these microbes made 100 or so gas vesicles per cell. They then injected the resulting smart probiotics into mice, either enema-style into the colon or directly into ovarian tumors, and found that they could reliably detect the location of the transgenic bugs inside the gut or the tumor by ultrasound—but not by light microscopy.
Although they have yet to try the technique in an animal larger than a mouse—let alone in a human—“we’ve tested it with several different frequencies of ultrasound,” says Shapiro, and shown that it works at the lower frequencies typically used clinically to penetrate deep tissues.
“This approach is very promising,” says Paul Miller, chief scientific officer at Synlogic, a startup cofounded by Collins to develop therapeutic microbes. “This may or may not ever end up being something that’s part of a candidate that goes into humans,” he notes, citing the additional metabolic burden imposed by the microbe having to produce the gas vesicles. But it could prove invaluable for studying the kinetics of designer microbes in proof-of-concept animal studies.
“We’re always looking for tools that can help us strengthen our ability to more confidently forecast how well something is going to work in humans from our preclinical data,” Miller says.
Ultrasound tracking of engineered bacteria might find a niche in clinical development as well. For example, this kind of tool might have helped a company called Vion Pharmaceuticals when, 18 years ago, it launched the only clinical trial to date involving a Salmonella strain to treat metastatic cancer.
At the time, the only way to tell whether the infused bacteria were successfully colonizing patient tumors was through tissue biopsies. Those biopsies, however, gave a mixed message about whether the bacteria were preferentially targeting the tumors or not. No antitumor effects were seen either, and Vion abandoned the therapy without launching further trials.
For years, David Bermudes, a former Vion scientist now at California State University, Northridge, has wondered: Was that decision premature? “That lack of ability to accurately detect the bacteria may have clouded what occurred clinically,” he says. Now, with the new ultrasound imaging technique, Bermudes could be in a better position to find out.
Additional diagnostic information might be gleaned if the ultrasound method was combined with other sound-based imaging modalities such as photoacoustic or thermoacoustic imaging. These techniques involve delivering light or radio-frequency pulses to the target tissue. Those pulses get converted to heat, causing a thermal expansion in the tissue and generating ultrasonic sound waves that can be analyzed to create detailed pictures of the body’s internal anatomy.
By merging these technologies, a clinician might simultaneously determine the precise location of a therapeutic microbe and image the surrounding tissue to see whether cells along the gut lining are inflamed, or whether a patient’s tumor is infiltrated with blood vessels.
“We looked into the [frequency] ranges used in both methods and there’s an overlap,” says Ricard Solé, a synthetic biologist at Pompeu Fabra University in Barcelona who wrote an accompanying commentary on the new research. “Thus, it would be possible to map the location of both engineered bacteria within or around tumors and other features such as vascularization—something not relevant in the example discussed in the Nature paper, but probably relevant in the future.”
source: www.spectrum.ieee.org